Omics Tests
The Omics Tests page allows you to view the genomic tests that have been indexed and ingested into the LifeOmic Platform.
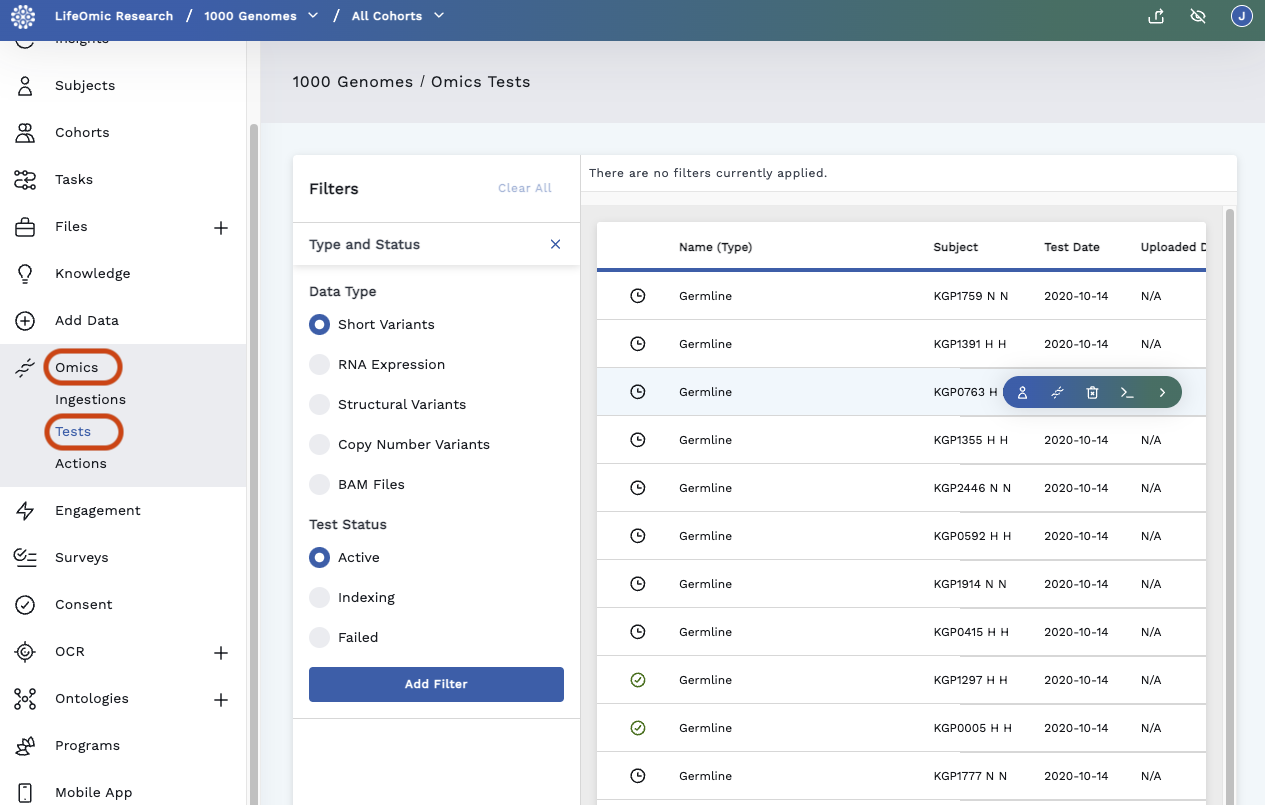
Filter and View Tests
The Omics Tests page allows you to view and filter the omics tests files.
-
On the left side panel, click Omics and Tests.
-
Under Filters, click the Plus icon to the right of the Type and Status box to display filter options.
-
Select desired filters and click Add Filter. The table filters the tests.
-
Hover on a test row to reveal a quick access menu:
- Open in Subject Viewer
- Open in Omics Explorer
- Open file under the [Files] tab(../files/index.md)
- Delete Test
- View CLI Help displays command to run in the LifeOmic CLI
- View Details takes you to the Test Detail page.
Delete Test requires deleteData>Access privileges. To grant privileges, see Access Control.
Clicking Delete Test only deletes the ingested data. This action cannot be undone. You can still find the base file used to generate the tests under the Files tab.
Omics Test Detail Page
Click a test row to open the Test Detail page. The Test Detail page shows all the variant and sequence types reported in the test in one Variant Sets table. It includes information about when the test was ingested, uploaded, and occurred.
The header of the page has the project name, the LifeOmic Platform tab, and the automated test file name. Beneath the header on the left is the test name and subject ID.
The top right has several quick access buttons. Depending on how your project is set up, all options may not be present.
- Open the subject in Subject Viewer
- Open the test in Omics Explorer
- View the test manifest
- Opens the file under the Files tab
- View CLI Help displays a command to run in the LifeOmic CLI
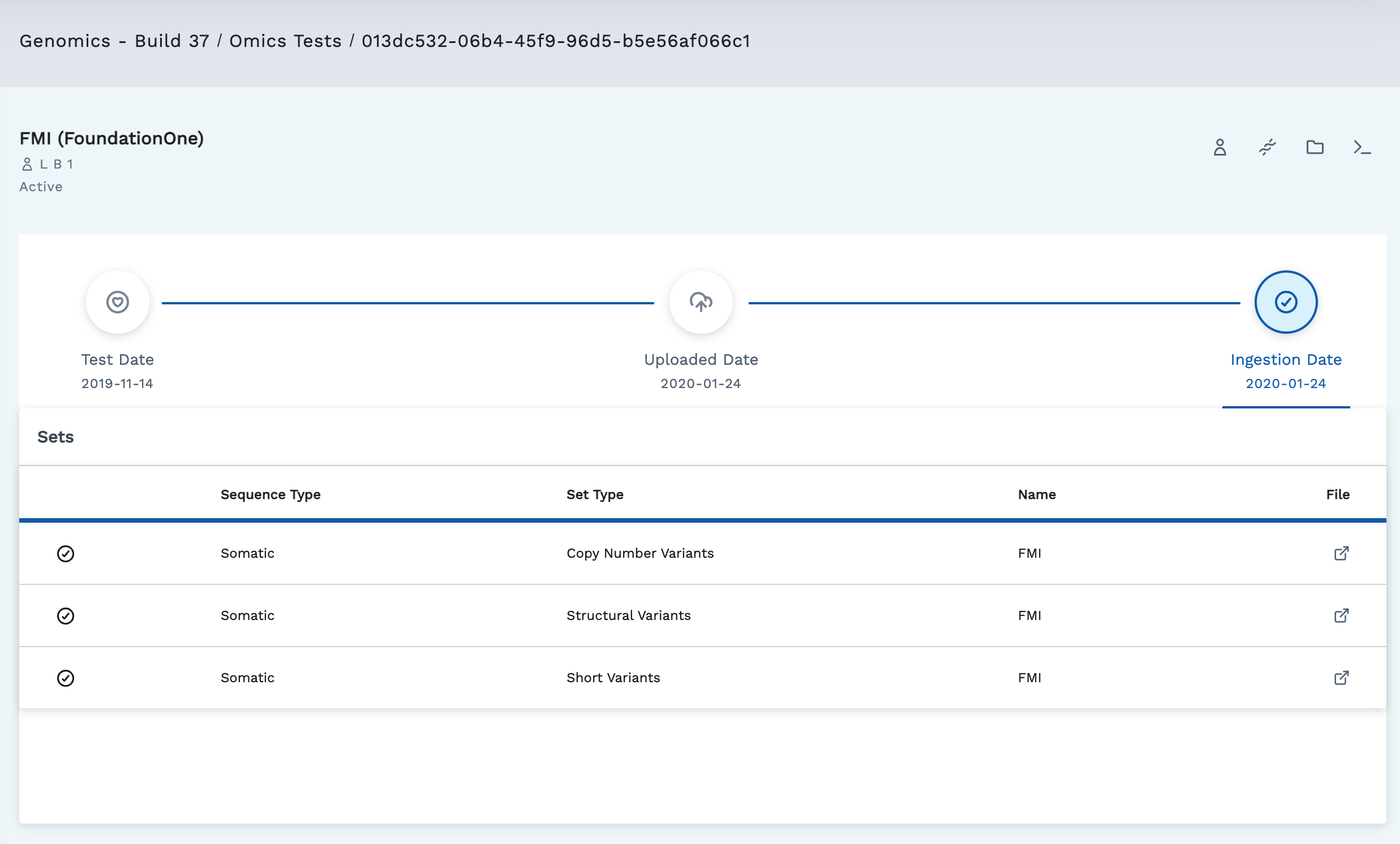
The Sets table shows all the variant & sequence types reported in the test. Table info includes:
- Test Status (active , indexing , or failed )
- Sequence Type (Somatic or Germline)
- Set Type (Copy Number Variant, Structural Variant, BAM Files, RNA Expression, and Short Variants)
- Name (Ashion, Ashion RNA, and others)
- File - Clicking this takes you to view the .vcf file and gives you the option to download the file in the CLI, web app, or share a link to the file.
If the .vcf file is too large, a warning displays and has an option to open the file.